Centromeres, located at the junction of chromosome arms, play an essential role in cell division: they ensure the equal distribution of the genome between the two daughter cells. Their activity depends in particular on a key protein: CENP-A (for CENtromeric Protein – A). Incorrect positioning of CENP-A can lead to an abnormal number of chromosomes in the daughter cells after division, a condition called aneuploidy, which is observed in many cancers.
Centromeres have always been difficult to study because they are made up of long, highly repetitive DNA sequences. However, the complete sequencing of the human genome (from telomere to telomere, T2T) completed in 2022 marked the beginning of a new era in centromere research. The complete centromere sequence revealed for the first time that CENP-A is concentrated in low-methylation[1] regions of centromeres, surrounded by highly methylated DNA. Scientists at Institut Curie sought to understand whether these methylation patterns influence the position and/or function of CENP-A.
Centromeric alterations linked to loss of DNA methylation
The Molecular Mechanisms of Chromosome Dynamics team, led by Dr. Daniele Fachinetti at Institut Curie (CNRS UMR 144 and 3664), developed cellular models to manipulate centromere methylation with great precision. The researchers showed that this epigenetic mark is essential for keeping CENP-A in place and ensuring proper centromere function. Thanks to the collaboration with Dr. Nicolas Altemose's team at Stanford University (United States) and the use of cutting-edge technology (DiMeLo-seq), the researchers were able to map the position of CENP-A and its links with methylation at unprecedented resolution. When centromeric DNA methylation is lost, CENP-A spreads into regions where it should not be present, potentially causing aneuploidy.
But that's not all: the researchers also showed that this centromeric DNA methylation independently controls the binding of a second protein, CENP-B, which—when too abundant— can make centromeric DNA more fragile and lead to errors in chromosome segregation.
Major consequences for genome stability
This work clearly demonstrates that this small chemical modification of DNA plays a crucial role in regulating the overall function of centromeres. It opens new perspectives for identifying other centromeric processes that may be directly or indirectly controlled by DNA methylation patterns.
These findings directly link epigenetic imbalances to chromosomal instability, which is observed in rare genetic disorders such as ICF syndromeas well as in many cancers. Importantly, the study also revealed that if methylation loss occurs gradually, some cells can adapt and survive—unlike when DNA methylation is lost too rapidly. This type of progressive change resembles what happens during cancer development.
"Our research shows for the first time that DNA methylation directly regulates centromere function. Because global DNA methylation loss is common in many cancers, these findings help us to better understand how it drives genomic instability by impairing centromere function" emphasizes Dr. Daniele Fachinetti.

Caption :
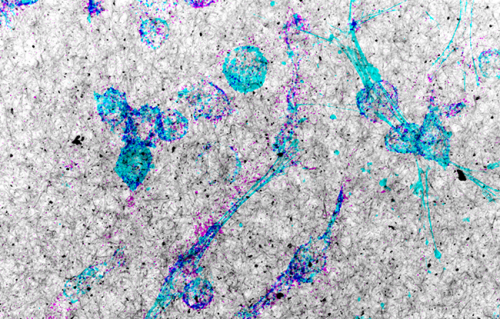
Top: Microscopic image of a normal chromosome with immunofluorescence staining for CENP-A (yellow) and CENP-B (magenta). Just below: Diagram showing highly methylated centromeric DNA (black circles) and highlighting the centromere dip region (CDR, green box), where DNA methylation is particularly low (white circles). The CDR coincides with the highest abundance of CENP-A.
Bottom: Chromosome after targeted centromere hypomethylation. The diagram, showing an enlarged view of the CDR, illustrates stronger immunofluorescence signals for CENP-A and CENP-B. This increase in centromeric protein levels alters centromere structure, leads to chromosome segregation errors, aneuploidy, and, in some cases, cell death.
© Catalina Salinas Luypaert
|
Référence : DNA methylation influences human centromere positioning and function - Catalina Salinas-Luypaert, Danilo Dubocanin, Rosa Jooyoung Lee, Lorena Andrade Ruiz, Riccardo Gamba, Marine Grison, Leonid Velikovsky, Annapaola Angrisani, Andrea Scelfo, Yuan Xu, Marie Dumont, Viviana Barra, Therese Wilhelm, Guillaume Velasco, Marialucrezia Losito, René Wardenaar, Claire Francastel, Floris Foijer, Geert J. P. L. Kops, Karen H. Miga, Nicolas Altemose & Daniele Fachinetti - Nature Genetics – 4 septembre 2025. DOI : https://doi.org/10.1038/s41588-025-02324-w |
[1] DNA methylation is an epigenetic modification consisting of the addition of chemical groups (CH3) to DNA. These do not alter its sequence but regulate its function.