Gene expression, protein transport, activation of signaling pathways, immune response, inter- and intra-cellular communication... All basic processes in biology are governed by complex mechanisms regulated by the physical proximity of molecules. How can we control and view these proximities to better understand and decipher the phenomena occurring in the cell? How can we exploit this proximity in a variety of applications?
 The teams of Prof. Arnaud Gautier, researcher at the Laboratory of Biomolecules (Sorbonne University, ENS-PSL University, CNRS) and Dr. Franck Perez, CNRS researcher director at Institut Curie (Cell Biology and Cancer unit/Institut Curie/CNRS) have designed an original and unique molecular tool capable of artificially controlling the proximity of two proteins in cells, but also of viewing their interactions in order to dissect the various molecular events involved.
The teams of Prof. Arnaud Gautier, researcher at the Laboratory of Biomolecules (Sorbonne University, ENS-PSL University, CNRS) and Dr. Franck Perez, CNRS researcher director at Institut Curie (Cell Biology and Cancer unit/Institut Curie/CNRS) have designed an original and unique molecular tool capable of artificially controlling the proximity of two proteins in cells, but also of viewing their interactions in order to dissect the various molecular events involved.
This new method has been named CATCHFIRE for Chemically Assisted Tethering of Chimera by Fluorogenic Induced Recognition.
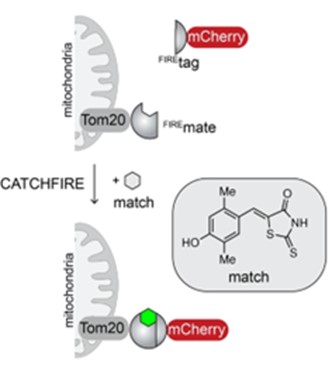
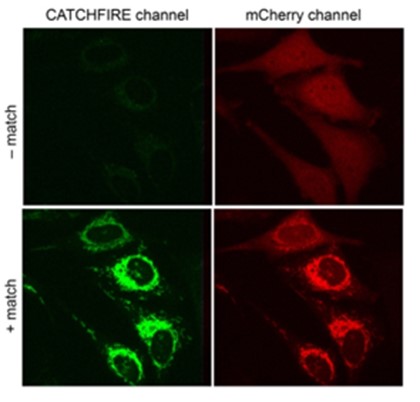
How does CATCHFIRE work? The two molecules in question (for example mCherry and Tom20 in the diagram opposite) are merged to two small protein domains (Firemate et Firetag), which are capable of interacting together in the presence of a small synthetic molecule, called “match”, acting as molecular adhesive.
When the two domains interact, the match molecule sees its fluorescence increase by a factor of 100. Researchers can then observe the newly-induced interaction via fluorescence microscopy. Another advantage of the system is that it’s reversible.
The advantage of CATCHFIRE is that this tool can reproduce itself and can be applied over and over with a number of proteins. This objective and quantitative approach has enabled researchers to control and track various interactions involved in the transport and location of proteins, secretory protein traffic, transport of organelles such as lysosomes and cellular mechanisms such as mitophagy.
In addition, they exploited the fluorogenic nature of CATCHFIRE to design new sensors - “biosensors” - that can quantify the activation of certain signaling pathways or trigger cellular processes such as apoptosis (programmed cell death).
 “Our approach elegantly illustrates how the combination of concepts and principles from two disciplines - chemistry and biology - enables us to design new and innovative molecular tools for the study and control of cell functions“ proclaim the authors, Dr. Franck Perez, biologist and CNRS researcher at Institut Curie and Prof. Arnaud Gautier, Professor of Chemistry at Sorbonne University. The application potential of CATCHFIRE is immense: the possibilities for studying basic biological processes are numerous, and the developments in biomedicine are promising, in particular in cellular therapy, which represents enormous hope for treatment of cancer and genetic and metabolic diseases, for example.”
“Our approach elegantly illustrates how the combination of concepts and principles from two disciplines - chemistry and biology - enables us to design new and innovative molecular tools for the study and control of cell functions“ proclaim the authors, Dr. Franck Perez, biologist and CNRS researcher at Institut Curie and Prof. Arnaud Gautier, Professor of Chemistry at Sorbonne University. The application potential of CATCHFIRE is immense: the possibilities for studying basic biological processes are numerous, and the developments in biomedicine are promising, in particular in cellular therapy, which represents enormous hope for treatment of cancer and genetic and metabolic diseases, for example.”
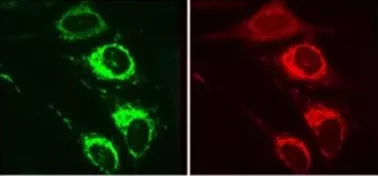
Legend : CATCHFIRE in action in mammal cells that jointly express the protein of the external membrane of Tom20 mitochondria merged with FIREmate and the red fluorescent protein mCherry merged with FIREtag. The addition of “match” induces the interaction of the two proteins (visible due to activation of its green fluorescence) leading to recruitment of mCherry at the surface of the mitochondria.
Reference: A fluorogenic chemically induced dimerization technology for controlling, imaging and sensing protein proximity. Sara Bottone, Octave Joliot, Zeyneb Vildan Cakil, Lina El Hajji, Louise-Marie Rakotoarison, Gaelle Boncompain, Franck Perez, Arnaud Gautier. Nature Methods, 28 August 2023 - https://doi.org/10.1038/s41592-023-01988-8